The transcriptome of each cell practically serves as a molecular fingerprint as it is unique and bundles all information that define cell identity, function, physiological status or sensitivity towards its environment. Single cell transcriptomics comprises the isolation and examination of transcriptomes of single cells/nuclei using RNA-sequencing, in situhybridization and the analysis of chromatin accessibility. Information from single cell/nucleus transcriptomes can be used for a range of research areas, including fundamental and applied plant biology. These include for example long-standing questions in evo-devo plant specialization and adaptation strategies to biotic and abiotic stimuli. The technology can be used to accelerate targeted plant breeding and genome engineering and precision fertilization. The Plant Single Cell platform at PSB offers the infrastructure to guide projects from the drawing board to comprehensive single cell datasets. Additionally, we continuously optimize protocols and de-risk novel single cell methods and technologies for various plant species. Publicly available scRNA-seq datasets from our platform can be accessed via our on-line browser tool.
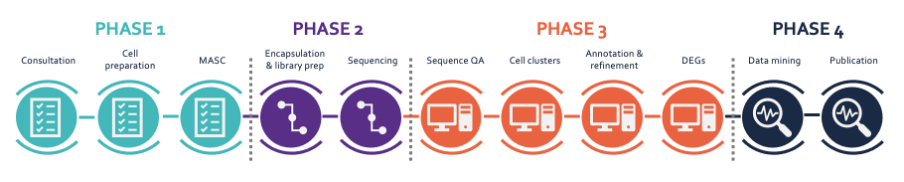
An end-to-end plant sc/snRNA-seq pipeline
We established a comprehensive pipeline that enables high-throughput processing of scRNA-seq and snRNA-seq samples from model plant species (e.g. Arabidopsis thaliana) and crops (e.g. maize, rice, soybean). Sample processing, including cell/nucleus isolation, cell/nucleus enrichment, sequencing and analysis, is done entirely within VIB using available core facilities. An advanced in-house bioinformatics pipeline for plant samples on multiple platforms (10X, BD) has also been established, allowing rapid and robust data processing, cell clustering, cluster annotation and visualization, and the analysis of developmental trajectories.
Technology implementation
Advanced methods and technologies for single cell/nucleus analyses develop very fast. We are continuously extending the portfolio of methods and technologies to study plant processes at a single cell/nucleus level. Recent additions to our portfolio include robust sc/snRNA-seq pipelines for multiple tissues and species, tissue fixation, long read sequencing, gentle sorting and enrichment protocols and evaluation of spatial transcriptomics providers.
Technology benchmarking
Early access to upcoming technology enables us to be on top of technological innovations in collaboration with the VIB early access programs. We regularly perform benchmark studies before including novel single cell technology platforms into our portfolio and pipelines. Current larger scale benchmark studies using plant samples include commercially available sc/snRNA-seq platforms and spatial transcriptomics providers.
The team
The Plant Single Cell platform at PSB is hosted in the De Rybel lab and coordinated by dr. Freya Persyn (management and communication), Jolien De Block (wetlab experience and support), Jonah Nolf (wetlab experience and support) and dr. Thomas Eekhout (operator and computational analysis).

