Due to the presence of a cell wall, plant cells are fixed within their tissue context and cannot move relative to each other during development. Plants thus need to rely on directed cell elongation and cell division to generate a full three-dimensional (3D) structure. Intrinsic polarity cues and cellular communication provide spatial information to plant cells and establishes their position relative to the tissue context and the axis of growth. This framework allows cells to integrate available information and orient their divisions in such a way that structured growth becomes possible. Controlling cell division orientations relative to the tissue axis is therefore the fundamental basis for 3D growth. Plants utilise two main types of cell division to allow directional growth: anticlinal and periclinal cell divisions. Anticlinal cell divisions (AD) generate more cells within a certain cell file (perpendicular to the tissue axis) and are thus one of the main drivers for longitudinal growth in the mitotically active regions of the plant, called meristems. This division type is obviously not sufficient, as it would only generate filamentous structures. In order to create a 3D structure, plants use formative or periclinal divisions (PD) that generate additional cell files (parallel to the tissue axis). This results in radial growth and the formation of new organs. It is clear that a very precise control of cell division orientation is crucial to allow normal 3D growth to occur. Indeed, excessive activity of factors triggering PD in plants result in strong radial expansion in for example root tissues. This illustrates the need for cells to divide in a particular orientation at a precise moment in development. Therefore, understanding the mechanisms that control cell proliferation and cell division orientation is a key question in developmental biology.
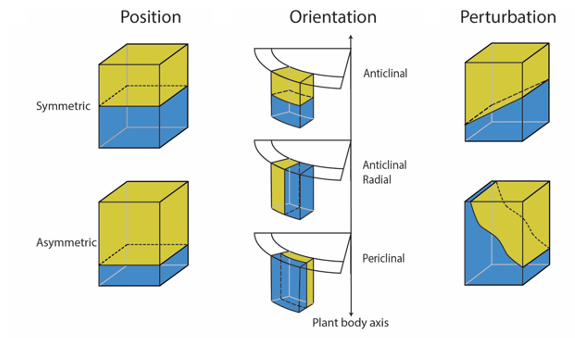
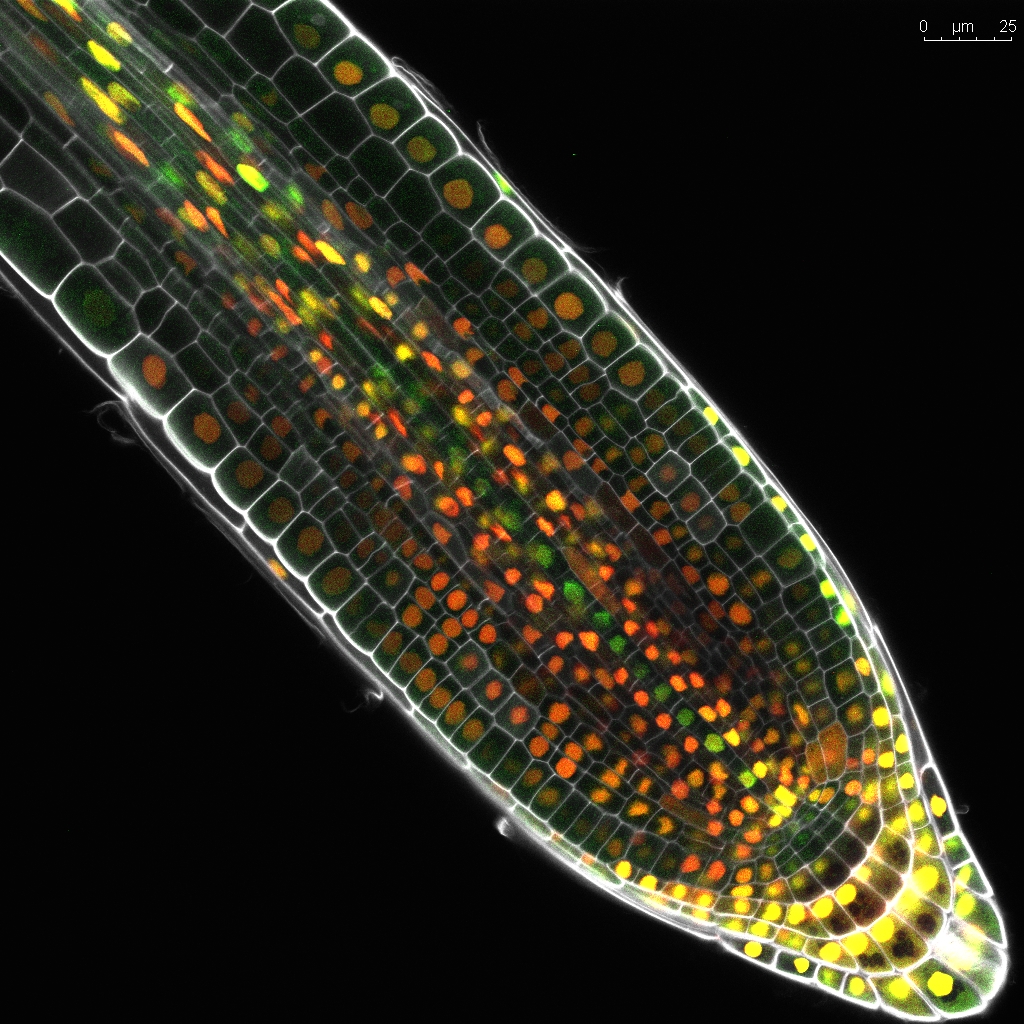
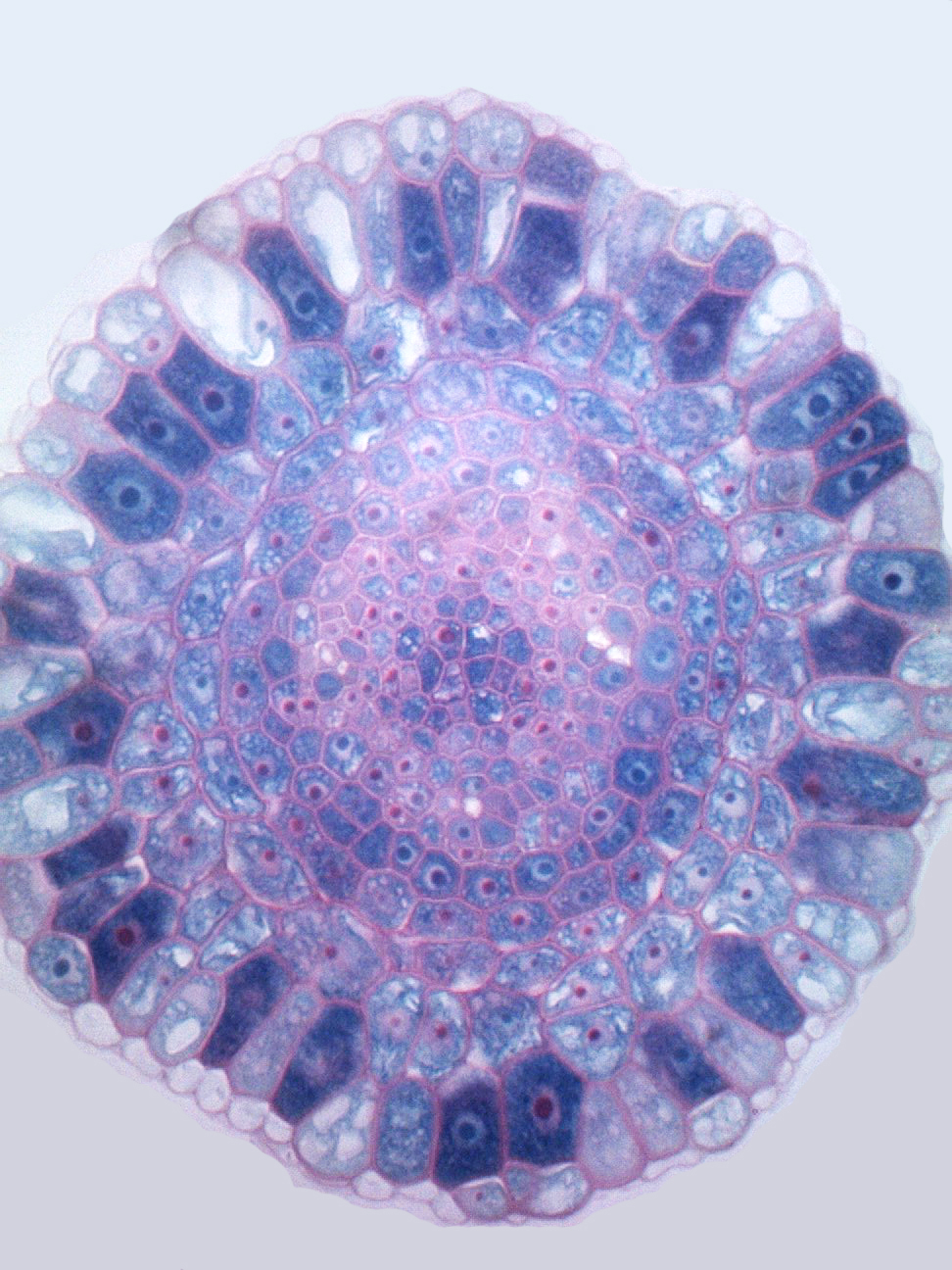
The Arabidopsis thaliana root is an excellent system to study oriented divisions, as there is clear spatial separation of morphologically distinct cell files along the main axis that are easy to follow throughout development. In particular the root tip, known as the root apical meristem, contains the stem cell niche and proliferation domains where new cells actively divide before reaching the elongation zone with their final cell fate. Oriented divisions are thus a key mechanism that shapes plant organs. Vascular tissues have the specific ability to undergo a tremendous amount of these oriented divisions and the additional cell files created this way generate almost all of the tissues that make up wood in trees, are crucial for source-to-sink transport throughout the plant and make up many edible structures such as fruits, roots and tubers. The plant vascular system is vital for transport of water, sugars and nutrients throughout the plant. Moreover, the evolution of an efficient fluid conducting system has allowed plants to grow beyond the size of non-vascular mosses. Despite the clear importance of this vascular proliferation for agricultural purposes and for our fundamental understanding of 3D growth, very little is known about how plant cells control the orientation of their cell divisions.
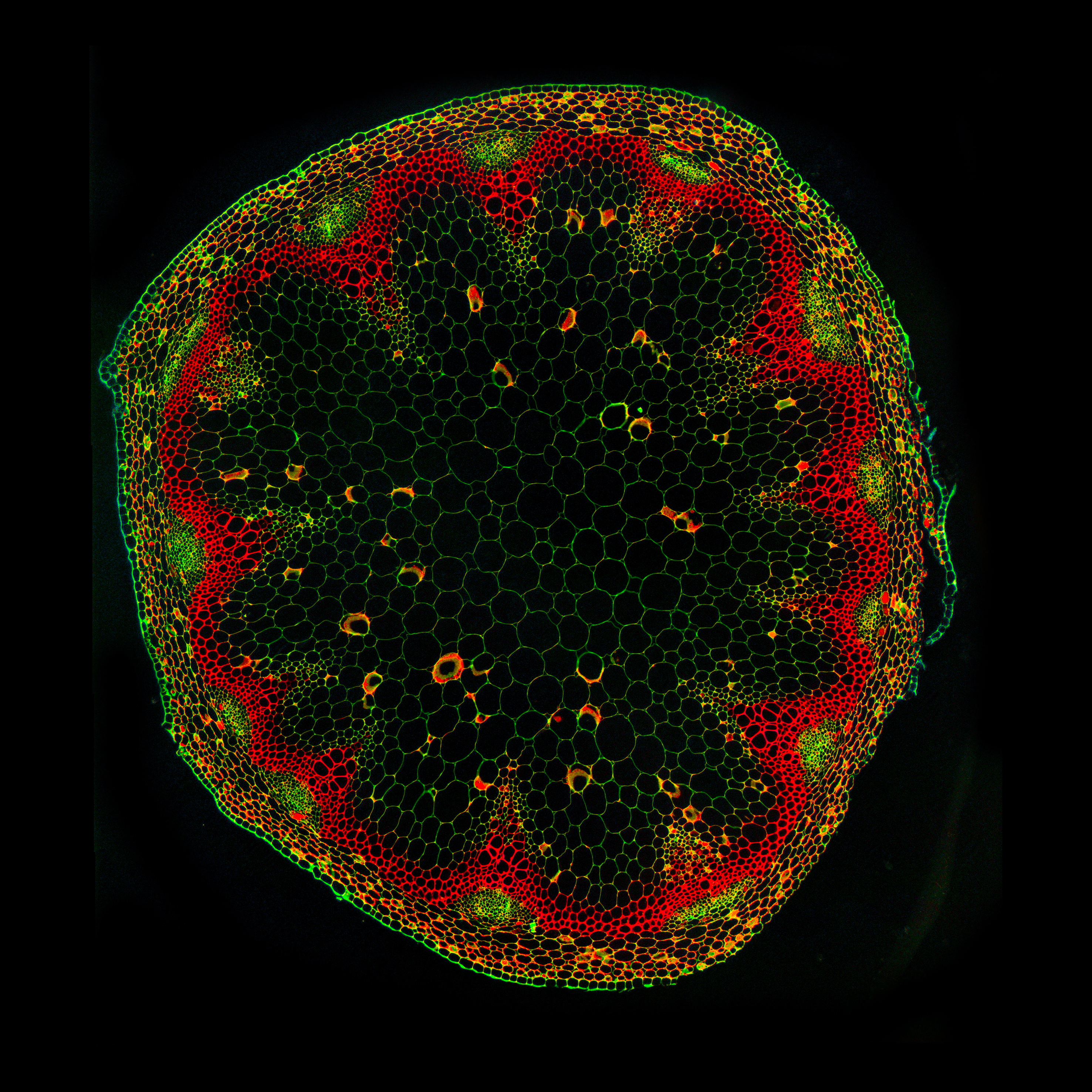
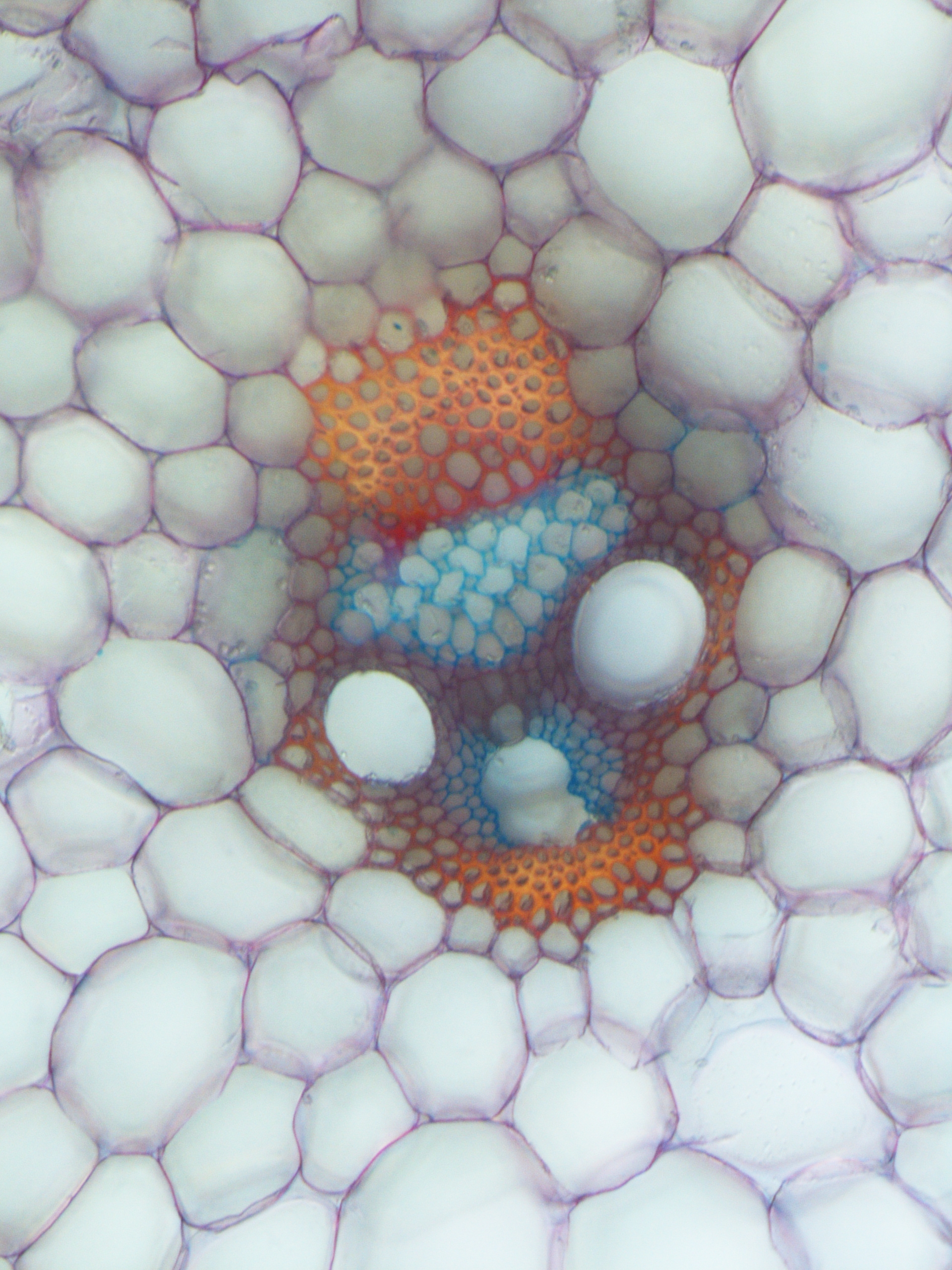
In the root, oriented divisions are of particular developmental importance, as they are essential for formation of a fully patterned vascular system: centralised xylem pole, procambium and the phloem poles towards the outer edge. For example, in the phloem lineage, a procambium cell divides periclinally, resulting in a sieve element precursor cell, which then after some rounds of periclinal/radial divisions generates proto-phloem, meta-phloem, and companion cells. This illustrates how oriented divisions are essential for both vascular proliferation and the formation of new cell identities. Understanding the mechanism of cell division orientation in the root vasculature would thus shed light not only on one of the open fundamental questions in developmental and cell biology but also allow a deeper understanding of tissue establishment, which would then be translated to technologies that guide vascular proliferation in crops.
We have previously identified and described the TMO5/LHW-DOF2.1 transcriptional pathway as a key regulator of vascular proliferation via controlling oriented divisions, particularly through induction of oriented cell divisions. Mechanistically, in the xylem, the bHLH transcription factors TMO5 and LHW dimerize and bind to the promoter regions of the cytokinin biosynthesis genes, LOG3, LOG4 and BGLU44. This locally produced cytokinin acts as a mobile signal, travelling to the procambium cells and inducing DOF2.1, a transcription factor that initiates oriented cell divisions without induction of other cytokinin dependent events. Loss-of-function tmo5/lhwor dof higher order mutants show strong root vascular defects, while ectopic misexpression of TMO5/LHW or DOF2.1 shows an increase of these cell divisions, irrespective of cell and tissue identity, suggesting that these factors are enough to control oriented cell divisions.
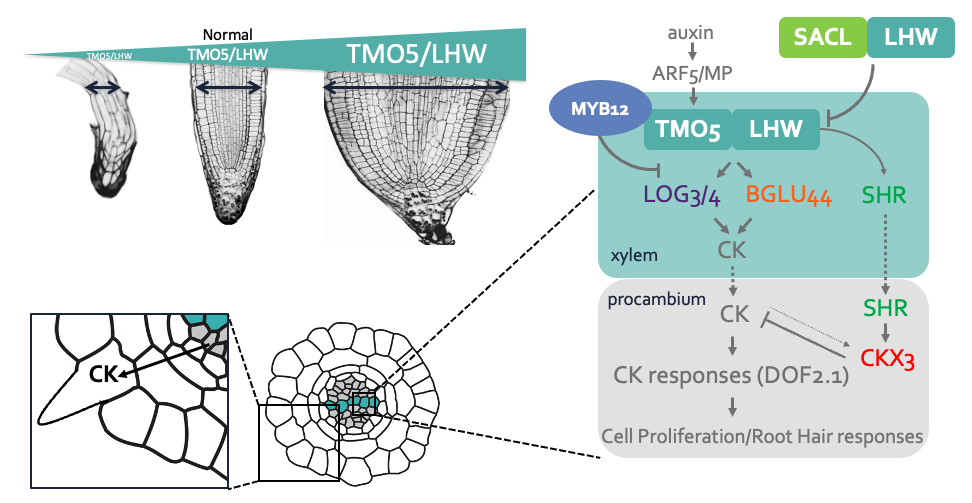
Figure: The TMO5/LHW pathway controls vascular cell proliferation and root hair responses to low phosphate conditions in the soil through local cytokinin biosynthesis and catabolism. Figure is based on the following publications from our group: Smet et al., Current Biology 2019; Wendrich et al., Science 2020; Yang et al., Nature Plants 2021; Mor, Pernisova et al., iScience 2022; Wybouw, Arents et al., J. Ex. Bot. 2023 and many great publications over the years from other research groups.
In summary, vascular tissue development depends on large and complex regulatory networks, whereby genetic, hormonal and environmental cues must be integrated and interpreted by the appropriate developmental output. We have been identifying and describing several transcriptional regulators controlling this process in recent years, but we are only at the beginning of uncovering the whole regulatory network responsible for vascular cell proliferation and cell division orientation. We tackle this research problem using both forward and reverse genetics approaches and combining different omics technologies including single cell/nucleus RNA-sequencing and chemical biology.
Work on this research topic is generously funded over the years by Ghent University (BOF18/PDO/151), the Research Foundation – Flanders (FWO grants G0D0515N and 1215820N), EMBO (EMBO ALTF 1005-2019) and the ERC (StG 714055 - TORPEDO and MSCA IF 885979 - DIVISION BELL).